Python TMVA
Python TMVA

Description
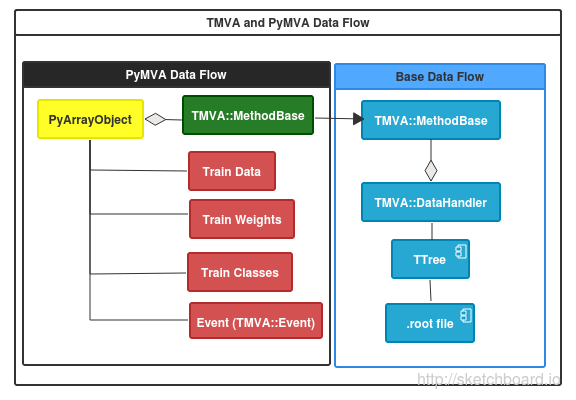
PyMVA is a set of plugins for TMVA package based on Python(SciKit learn) that consist in a set of classes that engage TMVA and allows new methods of classification and regression using by example SciKit’s modules.
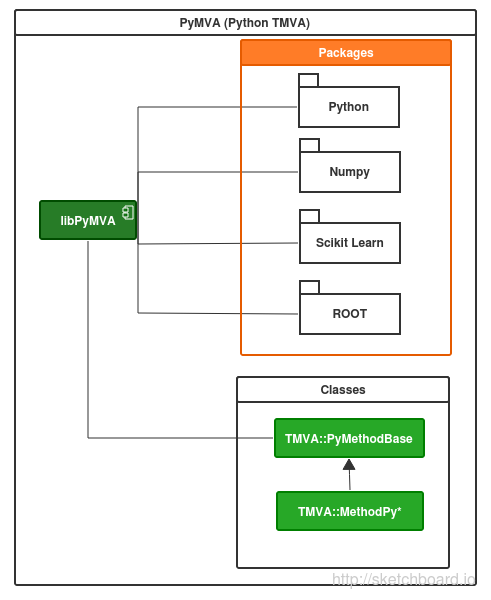
Installation
You can download the code from
git clone https://github.com/root-mirror/root.git
Install python libraries with headers and install the next package like a super user. You need to be sure that ROOT was compiled with python support in the same version odf that packages.
pip install scikit-learn
pip install numpy
PyRandomForest
This TMVA method was implemented using RandomForest Classifier Scikit Learn Random Forest
PyRandomForest Booking Options
| Option | Default Value | Predefined values | Description |
| NEstimators | 10 | - | integer, optional (default=10) . The number of trees in the forest. |
| Criterion | 'gini' | - | string, optional (default="gini"). The function to measure the quality of a split. Supported criteria are "gini" for the Gini impurity and "entropy" for the information gain. |
| MaxDepth | None | - | integer or None, optional (default=None).The maximum depth of the tree. If None, then nodes are expanded until all leaves are pure or until all leaves contain less than min_samples_split samples. Ignored if ``max_leaf_nodes`` is not None. |
| MinSamplesSplit | 2 | - | The minimum number of samples required to split an internal node. |
| MinSamplesLeaf | 1 | - | integer, optional (default=1). The minimum number of samples in newly created leaves. A split is discarded if after the split, one of the leaves would contain less then ``min_samples_leaf`` samples. |
| MinWeightFractionLeaf | 0 | - | float, optional (default=0.). The minimum weighted fraction of the input samples required to be at a leaf node. |
| MaxFeatures | 'auto' | - | int, float, string or None, optional (default="auto"). The number of features to consider when looking for the best split: - If int, then consider `max_features` features at each split. - If float, then `max_features` is a percentage and `int(max_features * n_features)` features are considered at each split. - If "auto", then `max_features=sqrt(n_features)`. - If "sqrt", then `max_features=sqrt(n_features)`. - If "log2", then `max_features=log2(n_features)`. - If None, then `max_features=n_features`. Note: the search for a split does not stop until at least one valid partition of the node samples is found, even if it requires to effectively inspect more than ``max_features`` features. |
| MaxLeafNodes | None | - | int or None, optional (default=None) Grow trees with ``max_leaf_nodes`` in best-first fashion. Best nodes are defined as relative reduction in impurity. If None then unlimited number of leaf nodes. If not None then ``max_depth`` will be ignored. |
| Bootstrap | kTRUE | - | boolean, optional (default=True). Whether bootstrap samples are used when building trees. |
| OoBScore | kFALSE | - | bool. Whether to use out-of-bag samples to estimate the generalization error. |
| NJobs | 1 | - | integer, optional (default=1). The number of jobs to run in parallel for both `fit` and `predict`. If -1, then the number of jobs is set to the number of cores. |
| RandomState | None | - | int, RandomState instance or None, optional (default=None). If int, random_state is the seed used by the random number generator; If RandomState instance, random_state is the random number generator;If None, the random number generator is the RandomState instance usedby `np.random`. |
| Verbose | 0 | - | int, optional (default=0). Controls the verbosity of the tree building process. |
| WarmStart | kFALSE | - | bool, optional (default=False). When set to ``True``, reuse the solution of the previous call to fit and add more estimators to the ensemble, otherwise, just fit a whole new forest. |
| ClassWeight | None | - | dict, list of dicts, "auto", "subsample" or None, optional. Weights associated with classes in the form ``{class_label: weight}``. If not given, all classes are supposed to have weight one. For multi-output problems, a list of dicts can be provided in the same order as the columns of y. The "auto" mode uses the values of y to automatically adjust weights inversely proportional to class frequencies in the input data. The "subsample" mode is the same as "auto" except that weights are computed based on the bootstrap sample for every tree grown. For multi-output, the weights of each column of y will be multiplied. Note that these weights wdict, list of dicts, "auto", "subsample" or None, optional Weights associated with classes in the form ``{class_label: weight}``. If not given, all classes are supposed to have weight one. For multi-output problems, a list of dicts can be provided in the same order as the columns of y. The "auto" mode uses the values of y to automatically adjust weights inversely proportional to class frequencies in the input data. The "subsample" mode is the same as "auto" except that weights are computed based on the bootstrap sample for every tree grown. For multi-output, the weights of each column of y will be multiplied. Note that these weights will be multiplied with sample_weight (passed through the fit method) if sample_weight is specified. ill be multiplied with sample_weight (passed through the fit method) if sample_weight is specified. |
PyRandomForest Booking Example
factory->BookMethod(dataloader,TMVA::Types::kPyRandomForest, "PyRandomForest","!V:NEstimators=200:Criterion=gini:MaxFeatures=auto:MaxDepth=6:MinSamplesLeaf=3:MinWeightFractionLeaf=0:Bootstrap=kTRUE" );
PyGTB (Gradient Trees Boosted)
This TMVA method was implemented using Gradient Trees Boosted Classifier Scikit Learn Gradient Trees Boosted
PyGTB (Gradient Trees Boosted)
This TMVA method was implemented using Gradient Trees Boosted Classifier
Scikit Learn Gradient Trees Boosted
PyGTB Booking Options
| Option | Default Value | Predefined values | Description |
| Loss | 'deviance' | - | {'deviance', 'exponential'}, optional (default='deviance'). loss function to be optimized. 'deviance' refers to deviance (= logistic regression) for classification with probabilistic outputs. For loss 'exponential' gradient boosting recovers the AdaBoost algorithm. |
| LearningRate | 0.1 | - | float, optional (default=0.1) learning rate shrinks the contribution of each tree by `learning_rate`. There is a trade-off between learning_rate and n_estimators. |
| NEstimators | 10 | - | integer, optional (default=10) . The number of trees in the forest. |
| Subsample | 1.0 | - | float, optional (default=1.0). The fraction of samples to be used for fitting the individual base learners. If smaller than 1.0 this results in Stochastic Gradient Boosting. `subsample` interacts with the parameter `n_estimators`. Choosing `subsample < 1.0` leads to a reduction of variance and an increase in bias. |
| MinSamplesSplit | 2 | - | The minimum number of samples required to split an internal node. |
| MinSamplesLeaf | 1 | - | integer, optional (default=1). The minimum number of samples in newly created leaves. A split is discarded if after the split, one of the leaves would contain less then ``min_samples_leaf`` samples. |
| MinWeightFractionLeaf | 0 | - | float, optional (default=0.). The minimum weighted fraction of the input samples required to be at a leaf node. |
| MaxDepth | 1 | - | integer , optional (default=3). maximum depth of the individual regression estimators. The maximum depth limits the number of nodes in the tree. Tune this parameter for best performance; the best value depends on the interaction of the input variables. Ignored if ``max_leaf_nodes`` is not None. |
| Init | None | - | BaseEstimator, None, optional (default=None) An estimator object that is used to compute the initial predictions. ``init`` has to provide ``fit`` and ``predict``. If None it uses ``loss.init_estimator``. |
| RandomState | None | - | int, RandomState instance or None, optional (default=None). If int, random_state is the seed used by the random number generator; If RandomState instance, random_state is the random number generator;If None, the random number generator is the RandomState instance usedby `np.random`. |
| MaxFeatures | None | - | int, float, string or None, optional (default=None). The number of features to consider when looking for the best split: - If int, then consider `max_features` features at each split. - If float, then `max_features` is a percentage and `int(max_features * n_features)` features are considered at each split. - If "auto", then `max_features=sqrt(n_features)`. - If "sqrt", then `max_features=sqrt(n_features)`. - If "log2", then `max_features=log2(n_features)`. - If None, then `max_features=n_features`. Choosing `max_features < n_features` leads to a reduction of variance and an increase in bias. Note: the search for a split does not stop until at least one valid partition of the node samples is found, even if it requires to effectively inspect more than ``max_features`` features. |
| MaxLeafNodes | None | - | int or None, optional (default=None). Grow trees with ``max_leaf_nodes`` in best-first fashion. Best nodes are defined as relative reduction in impurity. If None then unlimited number of leaf nodes. If not None then ``max_depth`` will be ignored. |
| WarmStart | kFALSE | - | bool, default: False. When set to ``True``, reuse the solution of the previous call to fit and add more estimators to the ensemble, otherwise, just erase the previous solution. |
PyGTB Booking Example
factory->BookMethod(dataloader,TMVA::Types::kPyGTB, "PyGTB","!V:NEstimators=150:Loss=deviance:LearningRate=0.1:Subsample=1:MaxDepth=3:MaxFeatures='auto'" );
PyAdaBoost
This TMVA method was implemented using AdaBoost Classifier Scikit Learn Adaptative Boost
PyAdaBoost Booking Options
| Option | Default Value | Predefined values | Description |
| BaseEstimator | None | - | object, optional (default=DecisionTreeClassifier). The base estimator from which the boosted ensemble is built. Support for sample weighting is required, as well as proper `classes_` and `n_classes_` attributes. |
| NEstimators | 50 | - | integer, optional (default=50).. The maximum number of estimators at which boosting is terminated.. In case of perfect fit, the learning procedure is stopped early. |
| LearningRate | 1.0 | - | loat, optional (default=1.). Learning rate shrinks the contribution of each classifier by ``learning_rate``. There is a trade-off between ``learning_rate`` and ``n_estimators``. |
| Algorithm | 'SAMME.R' | - | {'SAMME', 'SAMME.R'}, optional (default='SAMME.R'). If 'SAMME.R' then use the SAMME.R real boosting algorithm. ``base_estimator`` must support calculation of class probabilities. If 'SAMME' then use the SAMME discrete boosting algorithm. The SAMME.R algorithm typically converges faster than SAMME, achieving a lower test error with fewer boosting iterations. |
| RandomState | None | - | int, RandomState instance or None, optional (default=None). If int, random_state is the seed used by the random number generator; If RandomState instance, random_state is the random number generator; If None, the random number generator is the RandomState instance used by `np.random`. |
PyMVA Example
#include "TMVA/Factory.h"
#include "TMVA/Tools.h"
#include "TMVA/DataLoader.h"
#include "TMVA/MethodPyRandomForest.h"
#include "TMVA/MethodPyGTB.h"
#include "TMVA/MethodPyAdaBoost.h"
void Classification()
{
TMVA::Tools::Instance();
TString outfileName( "TMVA.root" );
TFile* outputFile = TFile::Open( outfileName, "RECREATE" );
TMVA::Factory *factory = new TMVA::Factory( "TMVAClassification", outputFile,
"!V:!Silent:Color:DrawProgressBar:Transformations=I;D;P;G,D:AnalysisType=Classification" );
TMVA::DataLoader dataloader("dl");
dataloader.AddVariable( "myvar1 := var1+var2", 'F' );
dataloader.AddVariable( "myvar2 := var1-var2", "Expression 2", "", 'F' );
dataloader.AddVariable( "var3", "Variable 3", "units", 'F' );
dataloader.AddVariable( "var4", "Variable 4", "units", 'F' );
dataloader.AddSpectator( "spec1 := var1*2", "Spectator 1", "units", 'F' );
dataloader.AddSpectator( "spec2 := var1*3", "Spectator 2", "units", 'F' );
TString fname = "./tmva_class_example.root";
if (gSystem->AccessPathName( fname )) // file does not exist in local directory
gSystem->Exec("curl -O http://root.cern.ch/files/tmva_class_example.root");
TFile *input = TFile::Open( fname );
std::cout << "--- TMVAClassification : Using input file: " << input->GetName() << std::endl;
// --- Register the training and test trees
TTree *tsignal = (TTree*)input->Get("TreeS");
TTree *tbackground = (TTree*)input->Get("TreeB");
// global event weights per tree (see below for setting event-wise weights)
Double_t signalWeight = 1.0;
Double_t backgroundWeight = 1.0;
// You can add an arbitrary number of signal or background trees
dataloader.AddSignalTree ( tsignal, signalWeight );
dataloader.AddBackgroundTree( tbackground, backgroundWeight );
// Set individual event weights (the variables must exist in the original TTree)
dataloader.SetBackgroundWeightExpression( "weight" );
// Apply additional cuts on the signal and background samples (can be different)
TCut mycuts = ""; // for example: TCut mycuts = "abs(var1)<0.5 && abs(var2-0.5)<1";
TCut mycutb = ""; // for example: TCut mycutb = "abs(var1)<0.5";
// Tell the factory how to use the training and testing events
dataloader.PrepareTrainingAndTestTree( mycuts, mycutb,
"nTrain_Signal=0:nTrain_Background=0:nTest_Signal=0:nTest_Background=0:SplitMode=Random:NormMode=NumEvents:!V" );
///////////////////
//Booking //
///////////////////
// PyMVA methods
factory->BookMethod(&dataloader, TMVA::Types::kPyRandomForest, "PyRandomForest",
"!V:NEstimators=100:Criterion=gini:MaxFeatures=auto:MaxDepth=6:MinSamplesLeaf=1:MinWeightFractionLeaf=0:Bootstrap=kTRUE" );
factory->BookMethod(&dataloader, TMVA::Types::kPyAdaBoost, "PyAdaBoost","!V:NEstimators=1000" );
factory->BookMethod(&dataloader, TMVA::Types::kPyGTB, "PyGTB","!V:NEstimators=150" );
// Train MVAs using the set of training events
factory->TrainAllMethods();
// ---- Evaluate all MVAs using the set of test events
factory->TestAllMethods();
// ----- Evaluate and compare performance of all configured MVAs
factory->EvaluateAllMethods();
// --------------------------------------------------------------
// Save the output
outputFile->Close();
std::cout << "==> Wrote root file: " << outputFile->GetName() << std::endl;
std::cout << "==> TMVAClassification is done!" << std::endl;
}
PyMVA Output
Evaluation results ranked by best signal efficiency and purity (area)
--------------------------------------------------------------------------------
MVA Signal efficiency at bkg eff.(error): | Sepa- Signifi-
Method: @B=0.01 @B=0.10 @B=0.30 ROC-integ. | ration: cance:
--------------------------------------------------------------------------------
PyGTB : 0.343(08) 0.751(07) 0.924(04) 0.914 | 0.539 1.514
PyAdaBoost : 0.275(08) 0.730(08) 0.914(05) 0.908 | 0.507 1.357
PyRandomForest : 0.233(07) 0.698(08) 0.903(05) 0.898 | 0.497 1.381
--------------------------------------------------------------------------------



